Research Highlights of All Cores in 2017
Core A
Core A of The CDAR Center has:
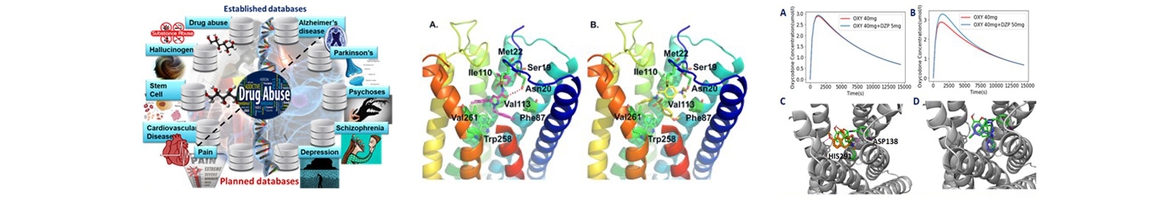
(1) established cloud computing drug-abuse and neurobiological diseases domain-specific databases, including hallucinogen (Xu et.al . J Mol Graph Model. 2016 Nov;70:284 ), cardiovascular disease (Zhang, et al. Sci Rep. 2016 Sep 28;6:33963) and stem cell (Zhang et.al. J Chem Inf Model. 2016 Oct 24;56(10)), in addition to its previously established drug abuse (Xie XQ et al, Front. Pharmacol. 2014 5:3),and Alzheimer’s disease (Liu et.al , J Chem Inf Model 2014;54(4):1050) knowledgebases. These platforms are integrated with online structure search function and data analysis tools to facilitate data-sharing and information exchange among scientific research communities for target/off-target identification neurobiological drug abuse research;
(2) implemented GPU-accelerated computational chemogenomics algorithms/tools for target identification to study the poly-pharmacophore and polyaddiction of abused drugs (Xue et.al. Acta Pharmacologica Sinica, 2017, in press; Wang et al, AAPS J. 2013 Apr;15(2):395);
(3) applied computational machine-/deep- learning computing chemogenomics algorithms/tools for design of novel small molecular probes for drug abuse and neurodisorders related research (Cha-Molstad et.al, Nat. Commun. 2017 Jul;8(1):102; Bian et.al. AAPS J. 2017 Jul;19(4):1235);
(4) utilized molecular systems pharmacology (MSP) techniques to explore molecular mechanisms associated with neurodisorder clinical outcomes (Hu et.al. Alzheimer & Dementia, under revision; Xue et al., Acta Pharmacologica Sinica, 2017);
(5) supported collaborations on drug abuse and neurodisorder related research. Along with Cores B and C, as well as outside collaborators, Core A has been working on CB2 signaling simulations, 𝛾-secretase ligand screening, and repurposing of current drugs for the treatment of drug abuse/addiction and neurodisorders (Wu, et. al. to be submitted to Bioinformatics; Zhang et.al, to be submitted to Molecular BioSystems; Hu et.al. Alzheimer & Dementia, under revision); and
(6) trained a new generation of drug abuse researchers.
Core B of The CDAR Center has accomplished/discovered the following:
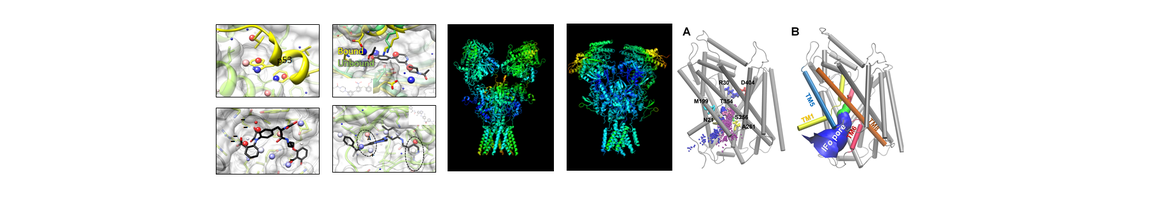
(1) Elucidation of anion conductance mechanism of human excitatory amino acid transporters and coupling to glutamate transport. In collaboration with Amara lab (NIH), they provide novel evidence on the molecular determinants of the dual function, substrate transport and anion permeation, of excitatory amino acid transporters (EAATs) (Cheng et al eLife 2017). This work opens new avenues toward illuminating how EAATs regulate synaptic transmission and neurological conditions and designing intervention methods.
(2) Allosteric modulation of dopamine transporter activity under conditions promoting its dimerization. They have examined the possible mechanisms of human dopamine transporter (hDAT) dimerization, as well as its dynamics using structure-based computations, site-directed mutagenesis, DA-uptake, and cross-linking (Dr. Garcia-Olivares and coworkers; NIH). Their study provides critical insights into approaches for examining the oligomerization of neurotransmitter transporters to shed light on their addictive drug modulation (Cheng, et al J. Biol. Chem. 2017). Furthermore, they have developed an efficient and robust anisotropic network model (ANM)-based methodology for modeling the intrinsic motions of proteins and their complexes and examined the collective dynamics of hDAT and Leucine transporter (Gur, et al J. Phys. Chem. B, 2017).
(3) Targeting of dopamine transporter to filopodia requires an outward-facing conformation of the transporter. Using quantitative live-cell fluorescence microscopy (Sorkin lab; U PITT) and molecular modeling (Bahar lab), they investigated the effects of the DAT inhibitor cocaine and its fluorescent analog JHC1-64 on the plasma membrane distribution of wild-type DAT and two DAT mutants. (Ma et al Sci. Rep. 2017).
(4) Development/upgrade of computational software/servers BalestraWeb, iGNM DB, DynOmics, and underlying computational methodology. Core B has upgraded their BalestraWeb server to improve the query search and overall performance. They have developed a new webserver DynOmics (Li et al Nucleic Acids Res. 2017) for efficient and accurate evaluation of the dynamics of structurally resolved systems in the context of their physiological environment.
(5) Other highlights: In collaboration with the Xie lab (Core A), they determined the mechanism of function of 𝛾-secretase, as well as its allosteric modulation or inhibition by substrate binding. They used to this aim as an extension of their membrane anisotropic network model (MembrANM) as well as druggability simulations. In collaboration with the Schreiber lab (Weizmann), they elucidated the regulation mechanism of interferon signaling (Li et a J Mol Biol 2017).
Core C
Core C of The CDAR Center has developed:
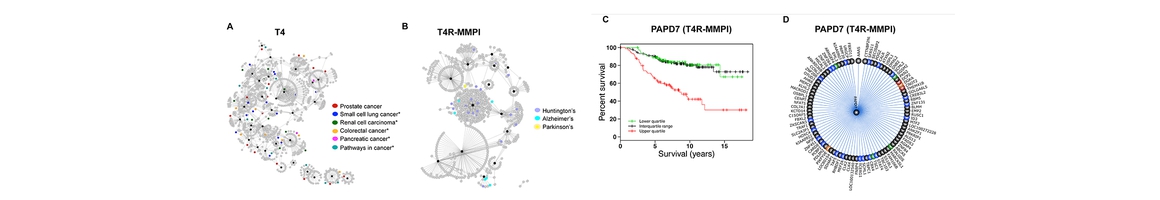
(1) A Time-Varying Group Sparse Additive Model (TV-GroupSpAM) for Genome-Wide Association Studies of Dynamic Complex Traits. In order to identify SNPs associated with the dynamic traits data collected from the Center for Education and Drug Abuse Research (CEDAR) at U Pitt, they have developed a new method, TV-GroupSpAM, for association mapping that considers dynamic phenotypes measured at a sequence of time points. They show that TV-GroupSpAM exhibits desirable empirical advantages over baseline methods on simulated datasets. They plan to use TV-GroupSpAM to identify DA-associated SNPs using the dynamic traits data from CEDAR.
(2) Discovering Weaker Genetic Associations with Validated Association, with Studies of Drug Abuse Disorder and Alzheimer’s Disease. Most existing methods barely take into account known/validated disease-specific genetic variants, making undiscovered variants harder to be identified because the signals of these variants are likely to masked by those of known variants. Core C proposes a new approach, Linear Regression with Validated Association (LRVA) that discovers variants by utilizing the knowledge of known associations, making the variants with weaker signals easier to be identified. They have applied their method to Drug Abuse Disorders data set collected from CEDAR provided to them by Dr. Michael Vanyukov (PI of a FRP), and discovered nearly two dozen novel SNPs associated with drug abuse disorders. Literature search suggests that many of these SNPs reside in genes previously known to be involved with substance abuse.
(3) Deep learning-based subdivision approach for large scale macromolecules structure recovery from electron cryo tomograms. In an effort to foster new ideas for the Center’s grant renewal, Core C has branched out to a new domain which will potentially allow them to collaborate more with Core B in the renewal phase. Cellular Electron CryoTomography is a powerful tool for analyzing 3D structures of macro-molecular complexes and their spatial organizations inside single cells. Due to the high degree of structural complexity, which makes de novo discovery of structures within cells challenging, they have proposed a new deep learning-based approach for subdividing subtomograms. Compared with existing methods, their approach achieves significant improvements in both discrimination ability and scalability. It can also find new structural classes and structures that do not exist in training data.