- Wang L, Xie X-Q. Cancer genomics: opportunities for medicinal chemistry? Future medicinal chemistry. 2016;8(4):357-359. doi:10.4155/fmc.16.1. PubMed PMID: 26976526; PubMed Central PMCID: PMC4847433.
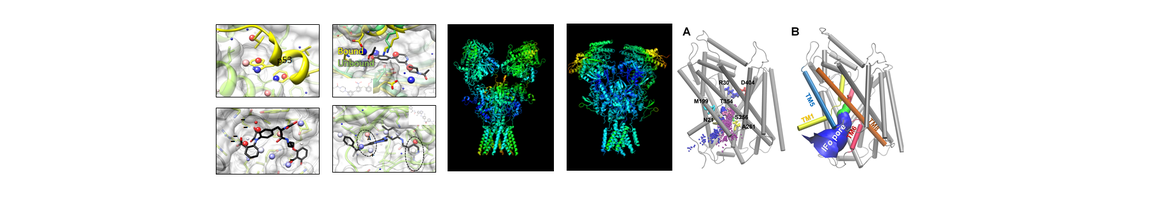
- Hu J, Feng Z, Ma S, Zhang Y, Tong Q, Alqarni MH, Gou X, Xie X-Q. Difference and influence of inactive and active states of cannabinoid receptor subtype CB2: from conformation to drug discovery. Journal of chemical information and modeling. 2016 May 26;56(6):1152-63. PubMed PMID: 27186994; PubMed Central PMCID: PMC5395206.
- Feng Z, Pearce LV, Zhang Y, Xing C, Herold BK, Ma S, Hu Z, Turcios NA, Yang P, Tong Q, McCall AK. Multi-functional diarylurea small molecule inhibitors of TRPV1 with therapeutic potential for neuroinflammation. The AAPS journal. 2016 Jul 1;18(4):898-913. PubMed PMID: 27000851; PubMed Central PMCID: PMC5333490.
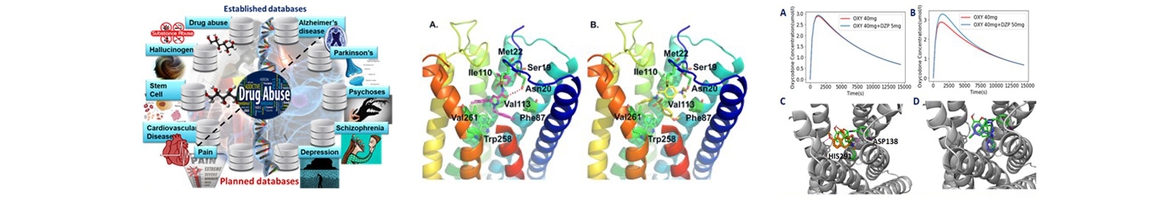
- Xu X, Ma S, Feng Z, Hu G, Wang L, Xie, X.-Q*. Chemogenomics knowledgebase and systems pharmacology for hallucinogen target identification-Salvinorin A as a case study. J Mol Graph Model. 2016 Nov;70:284-295. PMID: 27810775
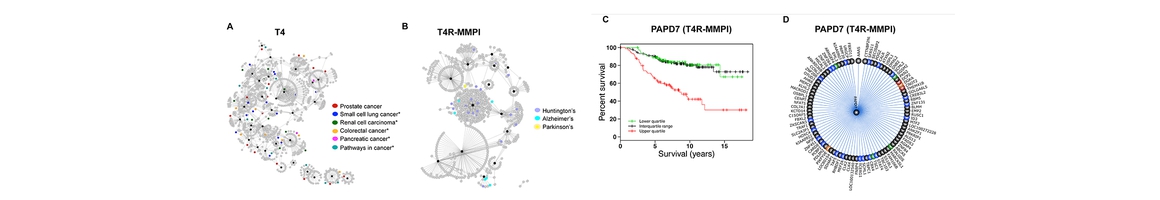
- Zhang H, Ma S, Feng Z, Wang D, Li C, Cao Y, Chen X, Liu A, Zhu Z, Zhang J, Zhang G, Chai Y, Wang L, Xie, X.-Q*. Cardiovascular Disease Chemogenomics Knowledgebase-guided Target Identification and Drug Synergy Mechanism Study of an Herbal Formula. Sci Rep. 2016 Sep 28;6:33963. PMID: 27678063
- Zhang Y, Wang L, Feng Z, Cheng H, McGuire TF, Ding Y, Cheng T, Gao Y, Xie, X.-Q*. StemCellCKB: An Integrated Stem Cell-Specific Chemogenomics KnowledgeBase for Target Identification and Systems-Pharmacology Research. J Chem Inf Model. 2016 Oct 24;56(10):1995-2004. PMID: 27643925
- Hu J, Hu Z, Zhang Y, Gou X, Mu Y, Wang L, Xie, X.-Q*. Metal binding mediated conformational change of XPA protein: a potential cytotoxic mechanism of nickel in the nucleotide excision repair. J Mol Model. 2016 Jul;22(7):156. PMID: 27307058
- Stern AM, Schurdak ME, Bahar I, Berg JM, Taylor DL. A perspective on implementing a quantitative systems pharmacology platform for drug discovery and the advancement of personalized medicine. Journal of biomolecular screening. 2016 Jul;21(6):521-34. PubMed PMID: 26962875; PubMed Central PMCID: PMC4917453.
- Liu B, Liu Q, Yang L, Palaniappan SK, Bahar I, Thiagarajan PS, Ding JL. Innate immune memory and homeostasis may be conferred through crosstalk between the TLR3 and TLR7 pathways. Sci. Signal.. 2016 Jul 12;9(436):ra70-. PubMed PMID: 27405980; PubMed Central PMCID: PMC5087126.
- Li H, Chang YY, Yang LW, Bahar I (2016) iGNM 2.0: the Gaussian network model database for bimolecular structural dynamics Nucleic Acids Res 44: D415-422 PMID: 26582920.
- Kurkcuoglu Z, Bahar I, Doruker P (2016) ClustENM: ENM-based sampling of essential conformational space at full atomic resolution. J. Chem. Theory Comput. 12:4549-62; PMID: 27494295.
- Jun I*, Cheng MH*, Sim E*, Jung J, Suh BL, Kim Y, Son H, Park K, Kim CH, Yoon J-H, Whitcomb DC, Bahar I, Lee MG (2016) Pore Dilation Increases the Bicarbonate Permeability of CFTR, ANO1, and Glycine Receptor Anion Channels. Journal of Physiology 594: 2929-55; PMID:26663196.
- Lee, S, Lozano, A., Kambadur, P., and Xing EP “An Efficient Nonlinear Regression Approach for Genome-wide Detection of Marginal and Interacting Genetic Variations.” Journal of Computational Biology 2016, 23(5): 372-389.
- Lee S, Kong S, Xing EP. “A Network-driven Approach for Genome-wide Association Mapping.” Bioinformatics 2016, 32(12):i164-i173. doi: 10.1093/bioinformatics/btw270.
- Bang, S. and Wu, W. “Naive-Bayes Ensemble: A New Approach to Classifying Unlabeled Multi-Class Asthma Subjects.” The IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2016).
- Marchetti-Bowick, J. Yin, J. Howrylak and E. P. Xing, A time-varying group sparse additive model for genome-wide association studies of dynamic complex traits, Bioinformatics 2016, 32 (19):btw347.
- Howrylak JA, Moll M, Weiss ST, Raby BA, Wu W, Xing EP. Gene Expression Profiling of Asthma Phenotypes Demonstrates Molecular Signatures of Atopy and Asthma Control. J Allergy Clin Immunol. 2016;137(5):1390-7.e6. Epub 2016/01/23. doi: 10.1016/j.jaci.2015.09.058. PubMed PMID: 26792209; PMCID: PMC4860055.